Abstract
The goal of this document is do introduce a reader to the current state of the field of genetic data compression, provide high level overview of the existing solutions on the market. As well as present our vision and current state of our project.
Who is this article written for?
This article might be useful for technical specialists or organization managers who are involved into genome sequencing projects, especially to those who are looking to optimize their data processing pipeline via more efficient genetic data compression.
Introduction to the problem of genetic data compression.
Genome sequencing raw data files, namely .fastq are huge. The size of one uncompressed file can vary from tens of GB to almost 1TB. A typical human whole genome sequencing experiment can produce 100s of GBs of data in FASTQ files with sequencing coverage often being 30x or higher. Which leads to the problem of disc space optimization for storage of those files.
General purpose compression tools
Most straight forward way to reduce file size is to use standard compression tools such as 7zip, gzip or pigz. Last one is actually parallel implementation of gzip, and probably is the fastest option among general purpose compression tool. In fact .fastq.gz file format is still industry standard for genetic data storage and many bioinformatic tools accept them as an input. But general purpose compression tools do not take into account the nature of .fasta/.fastq files, which are usually highly redundant on practice due to high coverage.
Genetic data specialized tools
To exploit this redundancy specialized compression tools have been developed. Those tools utilize various properties of genetic data files to make compression algorithms more efficient. First such specialized tools started to appear circa 2014 and keep evolving ever since.
Here you can see the graph of evolution of some open source solutions we were able to identify during our market research.
Blue - developed for short reads(NGS).
Green - developed for long reads(TGS).
Square - reference-free.
Rombus - reference-free and reference-based modes available.
Direction of arrow shows that authors of the paper claimed that their tool performed better than previous solution.
Solid line means that new solution was developed by the same team as previous one.
It is worth noting that tool represented on the graph above is not the complete list, there are more data compression projects, but some of them are obsolete( github wasn't update for 5+ years) and are not of great interest. The whole list of tools we were able to find during our research includes around 15 solutions, which you can observe at our compression tools overview list. We also put commercial solution into a separate category.
Commercial solutions
Most of them are closed source and requre paid subscription to use. But you can expect better performance metrics and developers support available.
For example you can have a look at Petagene or Genozip
Performance metrics and potential challenges.
There are several metrics should be kept in mind to choose the right solution for your needs.
Compression ratio.
Obviously compression ratio is the most important metric for compression algorithm. In case of .fastq file compression it may vary from 2.3 bit/base in case of general purpose algorithm like gzip up to 0.3 in case of highly specialized tools such as RENANO, CoLoRd, NanoSpring.
Compression speed.
Compression speed is also an important metric for evaluation of compression tools. The speed can depend on several factors such as: algorithm complexity, ability of parallelization, available hardware, especially sufficient amount of memory and number of CPU cores/threads available for parallel computation.
Speed vs CR tradeoff
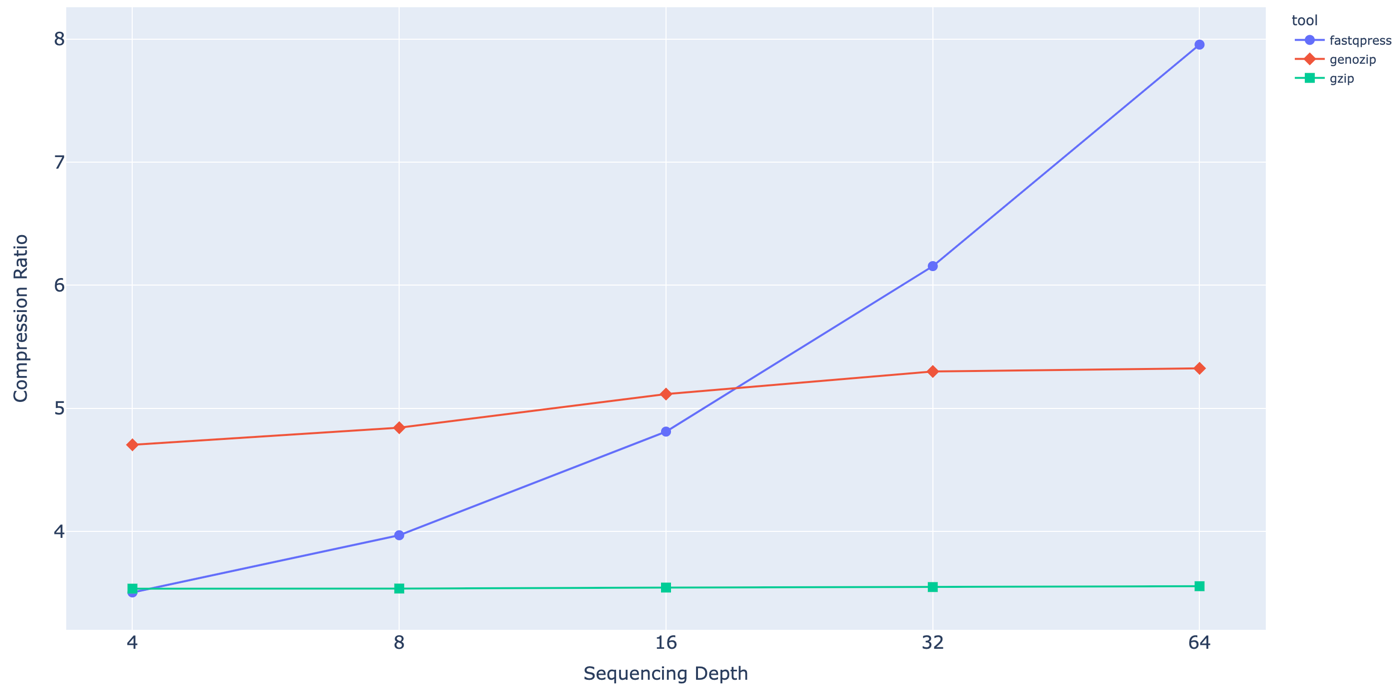
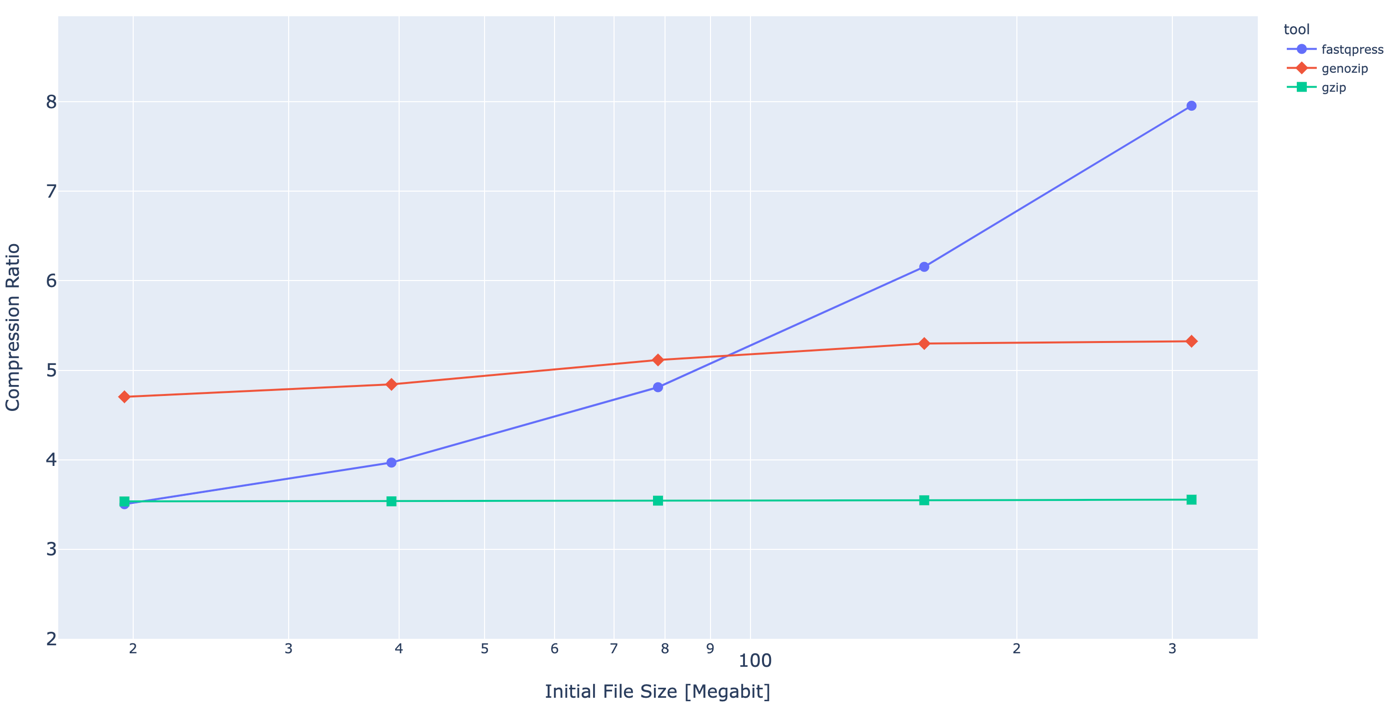
In case of some algorithms it could take up to 4 hours for human genome .fastq file compression. For example some opensource tools such as SPRING or CoLoRd are prioritizing optimization of compression ration in order to get closer to the theoretical limit which leads to poor speed performance. Whereas others are trying to find tradeoff between speed and CR. We think the good example of such tool is Genozip. This tools does not have such a good compression ratio as SPRING or CoLoRd, but it is still way better that general purpose compression algorithms and fast at the same time.
Other parameters to consider
-
Decompression speed.
Usually decompression requires less time and resources. -
Memory usage during compression and decompression 32GB usually should be enough.
-
Ability to deal with PacBio and Oxford NanoPore long reads. With appearance of a new sequencing hardware technologies such as PacBio and Oxford NanoPore, the reads length become significantly larger, 100k to 1kk base pairs per read. Which could be a problem for some compression tool, which were made for short read only.
-
Price One of the main reasons for data compression is storage use optimization. Everyone want to save some money. So the price is also an important factor to consider when choosing between compression solutions. Open source solution are free, but in many cases they lack proper support, so one might consider choosing among commercial solutions, but some of them might be more expansive then the others.
-
Open/close source. Licence. Code maintenance.
-
Reference free vs reference based.
-
Lossy/Lossless compression.
-
gzipped input support.
| Gzip | ColoRd | NanoSpring | RENANO | Genozip | Petagen | |
|---|---|---|---|---|---|---|
| Compression ratio | low | high | high | high | medium | No Data |
| Compression speed | medium | slow(hours) | slow(hours) | slow(hours) | fast(10th of minutes) | No Data |
| Decompression speed | high | low | low | low | high | No Data |
| Memory usage | ||||||
| Long read support | yes | yes | yes | yes | yes | yes |
| Price | Free | Free, but no changes allowed(GPL Licence) | Free | Free | $3500-$20000/Year | No data |
| Source code access | Open source | Open source | Open source | Open source | NOT AN OPEN SOURCE, but source code is available | No source code available |
| Licence type | GPLv3 | GPLv3 | MIT | MIT | Licence | Comercial |
| Lossy/Lossless compression | Lossless | Lossy and Lossless mode | Lossless | Lossless | Lossless | No data |
| gzipped input support | Not applicable | No | No | No | yes | No Data |
| VCF support | yes | No | No | No | yes | yes |
| BAM support | yes | No | No | No | yes | yes |
| Reference-free compression | yes | yes | yes | yes | yes | No Data |
| Reference-based compression | no | yes | yes | yes | yes | No Data |
Our vision.
We can see that field of genetic data compression is evolving and new solution appear every year. But from our perspective there is still a lot of room for improvement and competition. Especially in the dimension of finding right balance between execution speed and compression ratio.
Our current solution
For now we have a short read only compression tool, which has better CR and faster compression and decompression speed than current industry standard. Our approach is a combination of a simple preprocessing heuristics and usage of standard compression tools. Despite the fact that this approach is less efficient it term of compression ratio than such tools as ColoRd, NanoSpring or RENANO, We see this direction as most promising due to linear complexity of algorithm and its ability to utilize multiple cores. Which makes it way faster(minutes instead of hours) and still allows to keep compression ratio on the decent level.
Experiment
We have conducted comparison test on the small dataset between several tool, including our solution which we called fastqpress.
Experiment result data
| Tool | initial_size [byte] | result_size [byte] | time_to_compress [sec] | time_to_decompress [sec] | coverage | CR |
|---|---|---|---|---|---|---|
| fastqpress | 78544441 | 16327075 | 0.79 | 0.482 | 16 | 4.810686605 |
| fastqpress | 157185841 | 25535324 | 1.63 | 0.741 | 32 | 6.15562352 |
| fastqpress | 315420410 | 39648233 | 3.45 | 1.481 | 64 | 7.955472064 |
| genozip | 78544441 | 15354516 | 1.761 | 0.578 | 16 | 5.115396734 |
| genozip | 157185841 | 29662480 | 3.66 | 0.867 | 32 | 5.29914697 |
| genozip | 315420410 | 59240011 | 5.778 | 1.36 | 64 | 5.324448876 |
| colord | 78544441 | 7073538 | 6.384 | 1.262 | 16 | 11.10398234 |
| colord | 157185841 | 12443013 | 16.291 | 2.423 | 32 | 12.63245815 |
| colord | 315420410 | 22479791 | 41.947 | 4.185 | 64 | 14.03128748 |
| gzip | 78544441 | 22166272 | 10.104 | 0.383 | 16 | 3.543421329 |
| gzip | 157185841 | 44308116 | 20.281 | 0.768 | 32 | 3.547563182 |
| gzip | 315420410 | 88743282 | 39.807 | 1.494 | 64 | 3.554301834 |
Dependency of Compression ration from sequencing coverage.
Dependency of Compression ration from initial file size.
More benchmarks
Collaboration proposal and feedback collection
We are still at the early stage of development and willing to prioritize those features which are most valuable to our potential customers. We value every bit of information from industry experts and would be happy to talk in deep details about your current issue in genomic data processing pipeline. So please feel free to contact us at isachenkoa@gmal.com or subscribe to our email update list.